Back in college, when I was fully indoctrinated in germ theory, I wrote a paper titled ‘Speculations on How Sexually Transmitted Diseases Might Induce Host Behavior Changes’. A lot of that research stayed with me, such as a disease they say is specific only to rabbits and hares, called myxomatosis. I cited this disease as an example of how a ‘virus’ evolves from harmful virulence to benignancy. So when I recently read about ‘myxomatosis’ and the ‘myxoma virus’ that supposedly causes it, I went down that rabbit warren as a specific deep dive, just like Sam Bailey did for the tobacco mosaic ‘virus’: https://drsambailey.com/resources/videos/viruses-unplugged/tobacco-mosaic-virus-the-beginning-and-end-of-virology/ , considered the very first ‘virus’ ‘discovered’.
Tobacco Mosaic ‘Virus’
Tracing back the history of research on the ‘tobacco mosaic virus’, Bailey details the flaws of nine papers on TMV: they did not run control experiments and did not show evidence of a ‘virus’ in the tobacco plants. In one experiment, they damaged the roots of the tobacco plants with wood or tools, as explained in Bailey’s video. There is no English version of the 1903 paper by its ‘discoverer’, Dmitri Ivanovsky. Here is the earliest paper cited, from 1898, by Martinus W. Beijerck, showing his thought process on how ‘infection’ occurs: https://www.apsnet.org/edcenter/apsnetfeatures/Documents/1998/BeijerckSpotDiseaseTobaccoLeaves.PDF
So they employ extreme methods that deplete the soil and damage the plants, and then assume ‘infection’, and then say this concludes that a ‘virus’ is the causative agent without showing what a ‘virus’ is. And then the virology textbooks use these citations as the foundations for ‘viruses’. The more general biology textbooks and popular books then use these citations and beliefs, and then everyone just assumes that this is the truth. But the lack of evidence is what Stefan Lanka showed when he won his German Higher Regional Court case when no scientific papers detailed the existence of a measles virus:
Myxomatosis in rabbits
Back to ‘myxomatosis’, I had even bought one of the books that I read for the research, called Evolution of Infectious Disease by Paul Ewald. I include this here as another example of how the author, the scientific community, and the layperson read these beliefs, incorporate them, and that’s all there is to it- who is going to investigate these assertions about mosquito vectors, rabbit species, what lethality means, what resistance means, etc.:
In the video below, at the 3:00 mark, the narrator says that a ‘virus known to kill rabbits’ was introduced to curb the population of rabbits in Australia starting around 1926, which is different from Ewald’s book, where he states the introduction was around the middle of the century. The video goes on to say that this ‘moxie’ or myxomatosis ‘virus’ has a large genome and the DNA ‘unusually, is double-stranded’. He says they could not infect rabbits in their natural habitat, only in labs. The warrens ‘infected with moxie would die off, but the problem was, it did not spread between those warrens’. The program was halted until after World War II, but then picked up again, where the moxie was introduced and killed 77 out of about 4000 rabbits. ‘By the end of July, not a single sick rabbit could be found.’ Then in 1951 they postulated that the higher amount of rains that year brought more mosquito vectors to spread the ‘virus’ more effectively. But then it says by 1995, the rabbits were ‘mostly immune to this version of the virus’. Then the narrator says moxie was replaced by a new virus called kelsovirus. I can’t find anything about this.
This example shows how when we delve into a topic, so many statements hinge on how they are interpreted. Where did they get these ‘virus’ stocks? How do they know they are actually viruses that cause a change in the DNA of the host rabbits? What does effectiveness mean- for example is 77 deaths out of 4000 ‘effective’?
The account of myxomatosis in What Really Makes You Ill
Dawn Lester and David Parker have done invaluable research for their book, What Really Makes You Ill. What they wrote regarding ‘myxomatosis’ is really compelling, and you will get a better understanding of what could be going on. Note what I have italicized:
The first attempt to introduce myxomatosis was conducted in 1926; it is reported to have been unsuccessful. Further attempts were made during the 1930s, but, although curtailed by the advent of WWII, they are also reported to have been unsuccessful. However, efforts to reduce the rabbit population by infecting them with myxomatosis were resumed in 1949 under a research programme headed by the Australian Commonwealth Scientific and Industrial Research Organisation (CSIRO).
…
“Several field trials failed, but in the Christmas-New Year period of 1950-51 the disease escaped from one of the four trial sites in the Murray valley and spread all over the Murray-Darling basin, killing millions of rabbits.”
This episode is claimed to have virtually eradicated the rabbit population and is regarded as the first ‘successful’ introduction of myxomatosis; Professor Fenner states that it was this ‘outbreak’ that inspired him to study the virology of the disease. He adds an extremely interesting comment that,
“The climatic conditions at the time of the outbreak of myxomatosis in the Murray-Darling Basin had been such that there was also an outbreak of encephalitis in that region…”
…
It is clear that the many efforts to introduce an ‘infectious disease’ of rabbits produced inconsistent results that are inexplicable from the perspective of the ‘germ theory’. These variable results are, however, more than adequately explicable from the perspective of toxic chemicals.
The phenomenal surge in rabbit numbers after 1859 had encouraged Australian farmers and landholders to utilise a variety of poisons in their attempts to exterminate the rabbit ‘pest’; they had therefore used poisons for almost a century before the myxomatosis programme began. An article entitled The Balance of Nature Upset in Australia on the Rewilding Australia website, refers to some of the poisons used for ‘pest control’ during the 19th century; they include strychnine, arsenic, cyanide and phosphorous baits. Lead arsenate is also documented to have been used as a pesticide, but, although not used to poison rabbits, this highly toxic substance is a persistent environmental contaminant.
The impression from most establishment reports about myxomatosis is that the rabbit ‘problem’ was not successfully tackled until the field studies of the 1950s. However, this would seem to be a false impression, as the Rewilding article cites a letter written in 1921 by an Australian who laments the loss of large animals that would have been natural predators of rabbits and other ‘pests’. The author of the letter claims that the loss of these predators was mainly the result of the ‘compulsory poison law’; he states, “We also have a compulsory poison law, which compels every landowner in the Commonwealth to continually poison his land, from year’s end to year’s end.” He also states that, as a consequence of this law, the land was, “…always covered more or less with the carcases of poisoned rabbits…”
…
Strangely, although poisons had been used over a significant period of time against both of these ‘pests’, there are no references to insecticides, pesticides or other chemicals in any of the seven papers of the field study series. Yet various types of pesticides had been used; as acknowledged by the Australian Department of the Environment website which refers to the introduction of organochlorine pesticides (OCPs) and states, “Since they were first introduced into Australia in the mid 1940s, OCPs have been used in many commercial products…”
The OCPs introduced into Australia include DDT, chlordane, dieldrin and heptachlor, all of which have since been banned, either in the 1970s or 1980s, due to their extreme toxicity. The timing of the introduction of these extremely toxic substances and the subsequent virtual extermination of the rabbit population in 1950/51 cannot be dismissed as unconnected.
Organophosphates (OPs) are another class of toxic chemicals used in Australia to combat many insect ‘pests’, including mosquitoes; these chemicals act on the nervous system and have been associated with encephalitis. The simultaneous ‘outbreaks’ of encephalitis and myxomatosis indicates a very strong likelihood that OP insecticides had been used in the area. The field studies were solely focused on the belief in a viral cause of myxomatosis and so they failed to investigate any possible toxicological aspects of the rabbit ‘disease’. They clearly failed to recognise the possibility that insecticides may have been contributory factors; especially insecticides containing neurotoxins.
Phosphorus compounds are different from organophosphates, although both are recognised to be toxic. As previously discussed, phosphorus had been used as fertiliser since the late 19th century, and the work of Dr Scobey indicated the existence of a connection between phosphorus and paralysis. Furthermore, the Rewilding article refers to the use of phosphorous baits in the battle to poison rabbits.
The symptoms of phosphorus poisoning in humans include eye irritation, respiratory tract problems, eye burns and skin burns; interestingly, the symptoms of myxomatosis are red runny eyes, high fever, swollen mucous membranes, swollen eyes, nose, mouth and ears, plus respiratory problems. These similarities are striking and strongly indicate that, despite physiological differences between humans and rabbits, phosphorus-based chemicals should also be considered as likely contributory factors for the rabbit ‘disease’ that is referred to as ‘myxomatosis’.
Another highly toxic pesticide is sodium fluoroacetate, which is also known as 1080. This compound is an organofluorine; its effectiveness as a rodenticide is claimed to have been reported in the early 1940s. It is also reported to have been introduced into ‘rabbit control programmes’ in the early 1950s, which indicates that it may not have contributed to the first devastating ‘outbreak’ during 1950, but is likely to have contributed to some of the later ‘outbreaks’.
The world is awakening to the control of the masses by a few elite families. In this case, we know that the elite controllers such as the Standard Oil Rockefellers have influenced the petroleum and pharmaceutical industries, where many pharmaceuticals are manufactured using petrochemicals. The chemical pesticide sprays mentioned by Lester and Parker are all part of this industrial complex (remember, Standard Oil of NY owns CBS, NBC is at 30 Rock., ABC is owned by Disney). The rise in petrochemicals inevitably lead to increased toxicity, so the controllers need something else to blame. Researchers who dare to point a finger at these chemicals will not get anywhere in the science complex controlled by this network.
The Complete DNA Sequence of Myxoma Virus
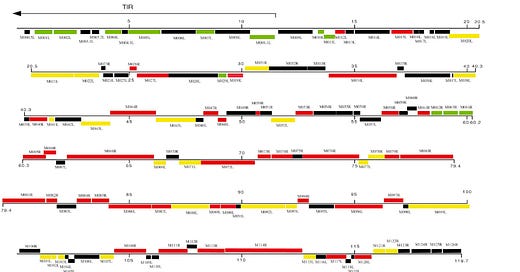
According to Wikipedia, myxoma is one of only four Leporipoxviruses. The others are for hares, squirrels, and another for rabbits, but these do not have Wikipedia entries. I guess all the other species similar to lagomorphs and rodents lucked out, or maybe scientists have not gotten around to discovering pox ‘viruses’ for them. Pox or pock marks of course implies that the body is trying to rid itself of toxins via the skin- like acne, warts, sweat, keloids, rashes, pus, inflammatory responses, eczema, leprosy, chicken pox, etc.: https://www.healthline.com/health/rashes#causes . Interesting, isn’t it, how they classify ‘viruses’ by function/morphology, where animals from different orders can fall prey to these ‘viruses’? And the MSW and MSD strains of the ‘California myxoma virus’, according to this paper, is 100% lethal to laboratory rabbits: https://www.sciencedirect.com/science/article/pii/S0042682205008056?via%3Dihub . They don’t state how many rabbits were tested, but how are these ‘viruses’ replicating without a living host?
I will now reference this paper, “The Complete DNA Sequence of Myxoma Virus.” The paper states that “the genomic DNA sequence of myxoma virus, strain Lausanne, was determined”, but that it was determined from analysing past DNA sequences:
Database searches revealed a central core of approximately 120 kb that encodes more than 100 genes that exhibit close relationships to the conserved genes of members of other poxvirus genera. Open reading frames with predicted signal sequences, localization motifs, or homology to known proteins with immunomodulatory or host-range functions were examined more extensively for predicted features such as hydrophobic regions, nucleic acid binding domains, ankyrin repeats, serpin signatures, lectin domains. and structural cysteine spacings. As a result, several novel, potentially immunomodulatory proteins have been identified…
Since this ‘complete DNA sequence’ paper is from 1999, could there be room for even more ‘novel’, ‘predicted’, and ‘potential’ sequences in the future, at least for this ‘Lausanne strain’, however they determine that? Or is this the final answer that replaced the previously best research on this strain from 1989? Of course the reason given for such evolution of different ‘strains’ and updates is because of:
the growing repertoire of virus genes that confer the unique capacity of each poxvirus family member to counter the immune responses of the infected host.
And how did they determine this counter-evolution?
The authors state that:
The myxoma virus genome (strain Lausanne) was determined to be 161774 nucleotides in length
but that this number is ‘deduced’ and is ‘consistent’ but ‘falls slightly short’ compared to a previous Lausanne strain sequence paper from 1989:
Our deduced genomic length falls slightly short of the predicted 163.6 6 0.2 kb length based on restriction enzyme mapping of myxoma virus, strain Lausanne (Russell and Robbins, 1989). We compared the fragments predicted from the deduced sequence with those observed in the publication of Russell and Robbins, and the restriction enzyme fragment profiles were consistent with the published map (not shown).
Unfortunately, you have to purchase a copy if you want to see the Russell and Robbins paper: https://www.sciencedirect.com/science/article/abs/pii/0042682289903620?via%3Dihub, as well as the Esposito et al. paper: https://www.sciencedirect.com/science/article/abs/pii/0166093481900367 . However, the 1999 paper outlines the original process from the Esposito paper- I include some of it here just to show that there is no way anyone could reproduce this experiment. Reproducibility is crucial in science and is the reason for including a materials and methods section in the first place. It goes without saying that another crucial step in science- a control experiment, was not performed. They only analyzed the sequence from the ‘Lausanne strain’ that they purchased from ATCC:
Briefly, roller bottles of baby green monkey kidney cells were infected with myxoma virus (Lausanne strain, from ATCC), viral cores were isolated with sucrose cushion centrifugation, and viral DNA was extracted with phenol-chloroform. Random genomic fragments of viral DNA were generated by sonication at 0°C. The sheared DNA was then ethanol precipitated, and blunt ends were generated using both T4 and Klenow polymerases. After phenol- chloroform extraction and ethanol precipitation, the DNA was separated on a 0.8% agarose gel and fragments of 1.5-3.0 kb were excised and purified. With a 3:1 ratio of insert to vector ends, the myxoma DNA fragments were then blunt end ligated into the EcoRV site of calf intestinal phosphatase-treated pBluescript II (KS1) vector (Stratagene) by incubation at 16°C overnight with T4 DNA ligase (GIBCO). The ligation mixture was then transformed into Escherichia coli ElectroMAX DH10B electrocompetent cells (GIBCO). Polymerase chain reaction (PCR), using T3 and T7 primers (Promega), was performed directly on positive colonies to identify clones containing inserts of the appropriate size for sequence analysis. Cycle sequencing reactions were performed at the DNA Sequence Facility at The John P. Robarts Research Institute, which uses the ABI377 DNA sequencer. Sequencing reactions were performed initially with M13F/R primers and subsequently with uniquely tailored primers, when additional sequencing information was required from a given clone.

You can purchase the ‘Lausanne strain’ from ATCC (American Type Culture Collection) for $756, as of this writing: https://www.atcc.org/products/vr-1829 . This of course is circular reasoning, where virologists start with an assumed ‘virus’ that they purchase, and then try to identify and sequence it. Below is the infamous example of this circularity, from the CDC, for the novel 2019 ‘coronavirus’: “since no quantified ‘virus’ isolates of 2019-nCov are currently available,” they conveniently used some ‘stock’ RNA with the highlighted GenBank accession number below- but how did they choose this ‘stock’ RNA?:
A very crucial piece of information is missing from the materials and methods section- the number of PCR cycles run:
Hopefully it wasn’t 45 cycles like the Christian Drosten WHO protocol: https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf?sfvrsn=a9ef618c_2 . Doubling something 45 times amplifies the thing to over 35 trillion (245 = 35,184,372,088,832). Each doubling has a drastic effect, so doubling 44 times would be half, or about 17 trillion. That’s why Kary Mullis said you can find almost anything in your body once you get to that level: https://odysee.com/@Gamzuletova:9/Kary-Mullis---You-can-find-almost-Anything-in-Anybody:2?r=8dVRRoTGwQdtgeFTtCbgSstbHy2iixzm .
The GeneCodes Sequencher 3.0 software they used in 1999 is outdated- their homepage lists version 5.4.6 now (surely this version is the last and cannot be improved upon!). And notice in the yellow highlight above that they decided to go with an 80% minimum match of contigs. Not sure what that exactly means, except that you can be off by 20% and still say it’s a match.
The virologists had to make many assumptions and calculations regarding ‘gene assignment’ based on the tendencies of other ‘poxvirus’ ‘genomes’, ‘as established by BLASTp scores’. The italics are mine, and show all of the subjective assumptions they had to make:
With all available reading frames, the number of possible myxoma virus ORFs encoding proteins of a minimum length of 50 aa was calculated to be 425. We based our gene assignment strategy on the fact that poxviruses tend to have a compact and efficiently organized genome, with the genes aligned closely one after another or slightly overlapping. Based on homologies to known poxvirus genes (as established by BLASTp scores) and comparison with the ORFs predicted in the recently sequenced SFV genome (Willer et al., 1999), we have assigned 159 myxoma ORFs predicted to be colinear expressed genes.
I’m sure the assumptions that the previous ‘poxvirus’ genomes they analyzed could never be improved upon- except by this paper! ORFs are open reading frames, the supposed translation of the code after reading a start codon and ending before a stop codon. I suppose a sentence in a language is an analogy to ORFs, with a capital letter after a period that usually tells you to read a new sentence, and a period that tells you to end the sentence, generally speaking.
More arbitrary naming (and ‘renaming’) ‘based loosely’ on grasshoppers (Melanoplus sanguinipes) and another ‘poxvirus’ that virologists call ‘Molluscum contagiosum virus’ (contagious and looks like molluscs?), as well as what virologists call ‘Semliki forest virus’ or SFV, another ‘leporipoxvirus’ that supposedly affects rabbits:
Our gene nomenclature was based loosely on those used for Melanoplus sanguinipes and M. contagiosum (Afonso et al., 1999; Senkevich et al., 1997) and was devised to correspond closely with that of the SFV genome (Willer et al., 1999). Beginning at the left-most end of the genome, the TIR genes were designated in a fashion consistent with the names used in previous articles and then continued in increasing numerical order from the left-most genes. Thus M-T1 was renamed M001R/L, M-T2 became M002R/L, and so on until M156R (with the exclusion of M145). Four notable exceptions to the numeric scheme are M000.5 R/L, which is an ORF found only in the termini of the myxoma genome; M-T3A and M-T3C, which became M003.1 R/L and M003.2 R/L; SERP-1, which became M008.1 R/L; and M153.1R, which is uniquely found in myxoma.
I think basing the gene nomenclature from grasshoppers was the key!
And this sentence was interesting:
the central 120 kb of the myxoma genome represents most of the essential genetic information common to all poxviruses, whereas the flanking 40 kb (approximately 15 kb at the left and 25 kb at the right) are enriched for genes that are more demonstrably specific for the Leporipoxvirus genus.
Is this always the case with life forms- whatever proteins are needed specific to a species are found on the ends of the genome, not the core? Of course, this would imply that the most basic or primitive functions are in the middle of strands, but I doubt that it would be so simple. I hope someone has done the experiments or analyses to show this.
And according to the authors, myxoma ‘virus’ is different from other poxviruses in that:
There is little evidence of wasted space, and in contrast to other poxviruses, there is no indication of noncoding DNA or significant stretches of repetitive DNA within the TIR, with the exception of nine tandem copies of a conserved nonamer/decamer between M002L/R and M003.1L/R. Although the TIR region of sequenced poxviruses is variable in length and sequence, the regions generally encode a small number of genes and include significant stretches of noncoding DNA. For example, there is almost 6 kb of noncoding DNA at the termini of vaccinia Ankara strain (Antoine et al., 1998), 3 kb in entomopoxvirus (M. sanguinipes) (Afonso et al., 1999), 3 kb in M. contagiosum virus (Senkevich et al., 1997), and 4 kb in vaccinia Copenhagen (Johnson et al., 1993). In contrast, there are contiguous ORFs right up to the termini in myxoma.

My video discussing some of the many assumptions and generalities with the 159 proteins of this myxoma ‘virus’:
BLAST (Basic Local Alignment Search Tool) is a searchable database on the NIH government website which tries to match a given nucleotide sequence with what has already been catalogued.
The NCBI accession number for this particular myxoma sequence is AF170726 (since updated to AF170726.2 : https://www.ncbi.nlm.nih.gov/nuccore/AF170726.2/ ).
When we look at protein or gene sequences in the NCBI website, we get many results from different species from various taxonomic categories. Biologists will say that one reason for this is because some proteins are required for basic functions across different organisms. This could very well be the case, but if so, why aren’t some of these proteins occurring in more species, not just a select number? And there seems to be many other basic proteins that are missing or not catalogued yet in many ‘virus’ sequences. And I notice the inverse, where there are very specific, nonfunctional, or unrelated ‘genes’ for proteins within ‘virus’ sequences. To me it’s analogous to saying a warehouse has all the parts needed for a specific car model, but you find common parts like different sized tires, different sized doors, some screws, some glass, but then there are also some but not all parts specific to this model, say the antenna that only fits the correct way. And then you find some unknown parts that do not fit any car, at least that anyone knows about (the black colored ORFS in the above Figure 1). You be the judge.
One last point: there is a lack of standardization in genomics, and the myxoma paper is no exception, as I have shown. From Mike Stone’s article, Limitations in Genome Sequencing Technology and Data Analysis, genomic sequencing:
is currently characterized by a lack of standard operating procedures, quality management/quality assurance specifications, proficiency testing systems and even less approved standards along with high cost and uncertainty of data quality.
In September 2020, after the creation of the “SARS-COV-2” genome and the explosion of genomes submitted to the GISAID database, China sought to finally address the lack of standardization in genomics by releasing the High-throughput Sequencer Standards. Even with this release, it is noted that the industry is only now moving toward a semblance of standardization. The first sequenced genome was said to have been done in 1977. This lack of standardization has permeated for nearly 50 years. How can any genome created through varying methods over decades with constantly evolving technologies and techniques ever be considered accurate, especially given that many genomes today relie (sic) on references to older genomes made with outdated and less “accurate” technology?
Here are other informative articles from Mike Stone detailing many issues regarding genomic sequencing: How Accurate and Reliable are Genomes and Genome Contamination: A Widespread Problem.
Like anything in life, when we zoom into the finer fractal details of a topic, we see that the news-bite understanding is not what it seems. We need to zoom in for ourselves, not rely on ‘experts’ to tell us to trust them. Then we can shift the narrative and the grip they have on us. The magic spell we are under constantly changes- for example, when Fast Food Nation, Supersize Me, Omnivore’s Dilemma, etc., came out, things slowly changed regarding fast food and diet in general. We are slowly understanding the ruse of virology, and more investigation is to come. Thanks for reading.
More resources and links:
I made this video of screenshots I saved in December of 2020, and it still holds up:
Rabies- another ‘virus’ that supposedly changes the victim’s behavior, but Brendan Murphy details how the evidence says otherwise:
Samantha Bailey video on the Tobacco Mosaic ‘Virus’: https://odysee.com/@drsambailey:c/tobacco-mosaic-virus-the-beginning-and-end-of-virology:8?r=8dVRRoTGwQdtgeFTtCbgSstbHy2iixzm
David Crowe interviews PCR expert Stephen Bustin:
Myxoma Virus and the Leporipoxviruses: An Evolutionary Paradigm: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4379559/
My highlights from the The Complete DNA Sequence of Myxoma Virus paper.









Excellent research! Yes, people need to learn to read journal articles which point out the flaws of the supposed "findings" reported by "experts" and "authorities! But, starting with a basic distrust of any of the sources along the line is a good position!